| Correspondence
Izuho Hatada
Biological Genome Resource Center, Institute for Molecular and Cellular Regulation,Gunma University
|
| Strategy for MIAMI |
|
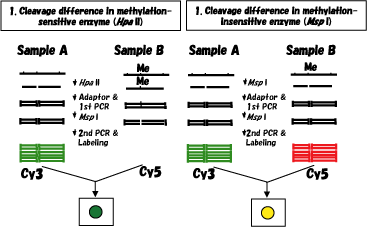
For the detection of cleavage difference in methylation-sensitive enzyme (Hpa II), which is associated with methylation difference, Hpa II digestion was followed by adaptor ligation and several cycles of PCR (1st PCR). At this stage, only DNA fragments that have methylated internal Hpa II sites before PCR retained Hpa II (Msp I) sites. Therefore, Msp I digestion and the following 2nd main PCR makes it impossible to amplify these methylated fragments. Amplified DNA fragments from two samples were labeled with Cy3 and Cy5, respectively, and co-hybridized to a microarray, and ratios of fluorescence intensities were used to calculate the methylation difference. On the other hand, for the detection of cleavage difference in methylation-insensitive enzyme (Msp I), which is used to judge the reliability of methylation difference, the same procedure is performed except for Msp I digestion at the first step. Cleavage differences in Hpa II and Msp I were used for analysis. If two samples have a restriction site polymorphism at an Hpa II site and no methylation difference, we should detect cleavage differences in both Hpa II and Msp I. Thus, we judge this case as having no methylation difference. If we observe a cleavage difference only in Hpa II, we can judge it as a true methylation difference. |