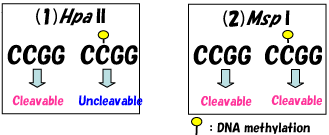
| Genome-wide profiling of DNA methylation has become important because epigenetic changes are thought to be involved in many diseases. We developed the first genome-wide DNA methylation profiling technology called Restriction Landmark Genomic Scanning (RLGS) (Ref. 1, 2) in 1991; however, this method cannot be applied to all genes in the genome and it requires some skill. We developed the Microarray-based Integrated Analysis of Methylation by Isoschizomers (MIAMI) method (Ref. 3) to resolve these problems. Restriction enzymes are used to detect DNA methylation in this method. It is well known that most DNA methylation is located at the C5 position of cytosine in a CpG (CG) sequence. A restriction enzyme called Hpa II recognizes and cleaves the CCGG sequence which includes CpG; however, it cannot cleave the same sequence when internal C is methylated (methylation-sensitive restriction enzyme). On the other hand, a restriction enzyme called Msp I, which recognizes the same sequence, cleaves both methylated and unmethylated CCGG sequences (methylation-insensitive restriction enzyme). With the MIAMI method, the difference in cleavage by both methylation-sensitive restriction enzyme Hpa II (1) , which is associated with methylation difference, and methylation-insensitive restriction enzyme Msp I (2) which is associated with difference without methylation was compared between samples. The reliability of the difference in Hpa II cleavage is judged using the difference in Msp I cleavage. If two samples have restriction site polymorphism at an Hpa II site and/or one of the samples has incomplete digestion at an Hpa II site, they will differ in cleavage to Hpa II without methylation difference. However, in this case, cleavage by methylation-insensitive Msp I at this site will also differ between samples, and can be judged as false positive.
|